General Info
NLRscape is an interactive atlas of plant NLRs equipped with a collection of easy, ready to use bioinformatic tools aimed for the exploration of the complex sequence landscape of this class of receptors in plants.
NLRscape is growing and input from users in this process is much welcomed. Currently the website is in its beta version - some functionalities are being optimized and might be on and off and subjected to changes or updates at some point.
The atlas aims to facilitate two types of use case:
- “Bottom-up” exploration by starting with a specific NLR sequence and further exploring its close-by homologs, sequence variability and other structural related characteristics.
- “Top-down” exploration by investigating the most representative NLR groups clustered according to homology, taxonomic spread or domain organisation.
Quick search
-
sequence name"zar1"
-
UKB ID"Q38834" (no spaces)
-
organism name"arabidopsis", "arabidopsis thaliana" (some species have synonym names already integrated, some don't)
-
full lineage"liliopsida", "magnoliopsida", "poales", any internal taxonomic node
-
domain exact organisation"CC-NBD-ARC1-ARC2-LRR", "X-NBD-ARC1-ARC2-LRR-KIN", etc
-
a particular domain or configuration"CC", "RPW8", "KIN", "NBD-ARC1-ARC2", "LRR-WRKY", etc
-
sequence"MVDAVVTVFLEKTLNILEEKGRTVSD" (parts of the sequence with exact match, no spaces / endline characters for the moment)
-
An improved search feature will be available soon, which will also allow blasting sequences for finding close but not identical matches as well.* all search fields are case insensitive.
Features
New features and updates of NLRscape are planned and will arrive in the following months..
Top-down
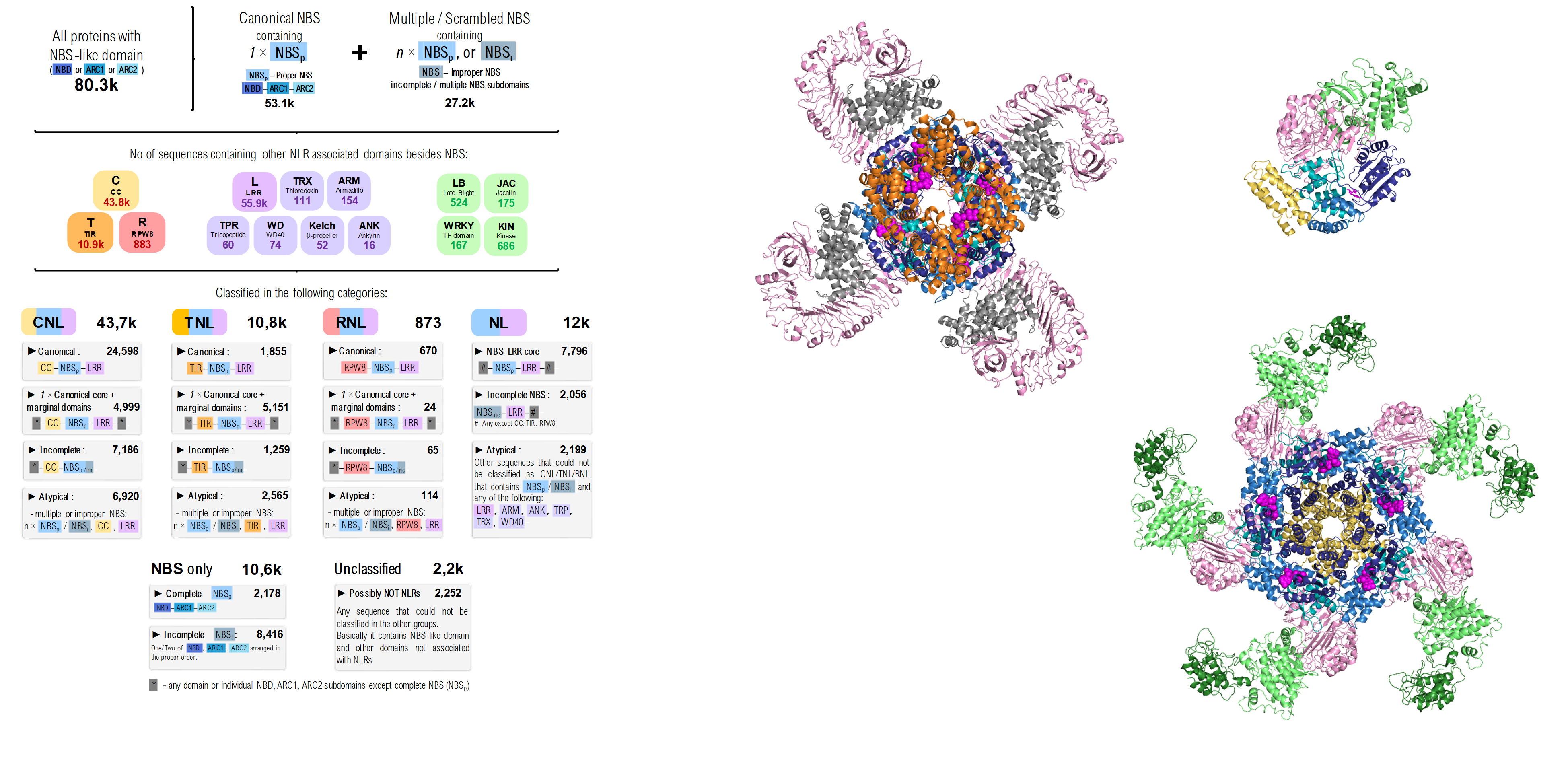
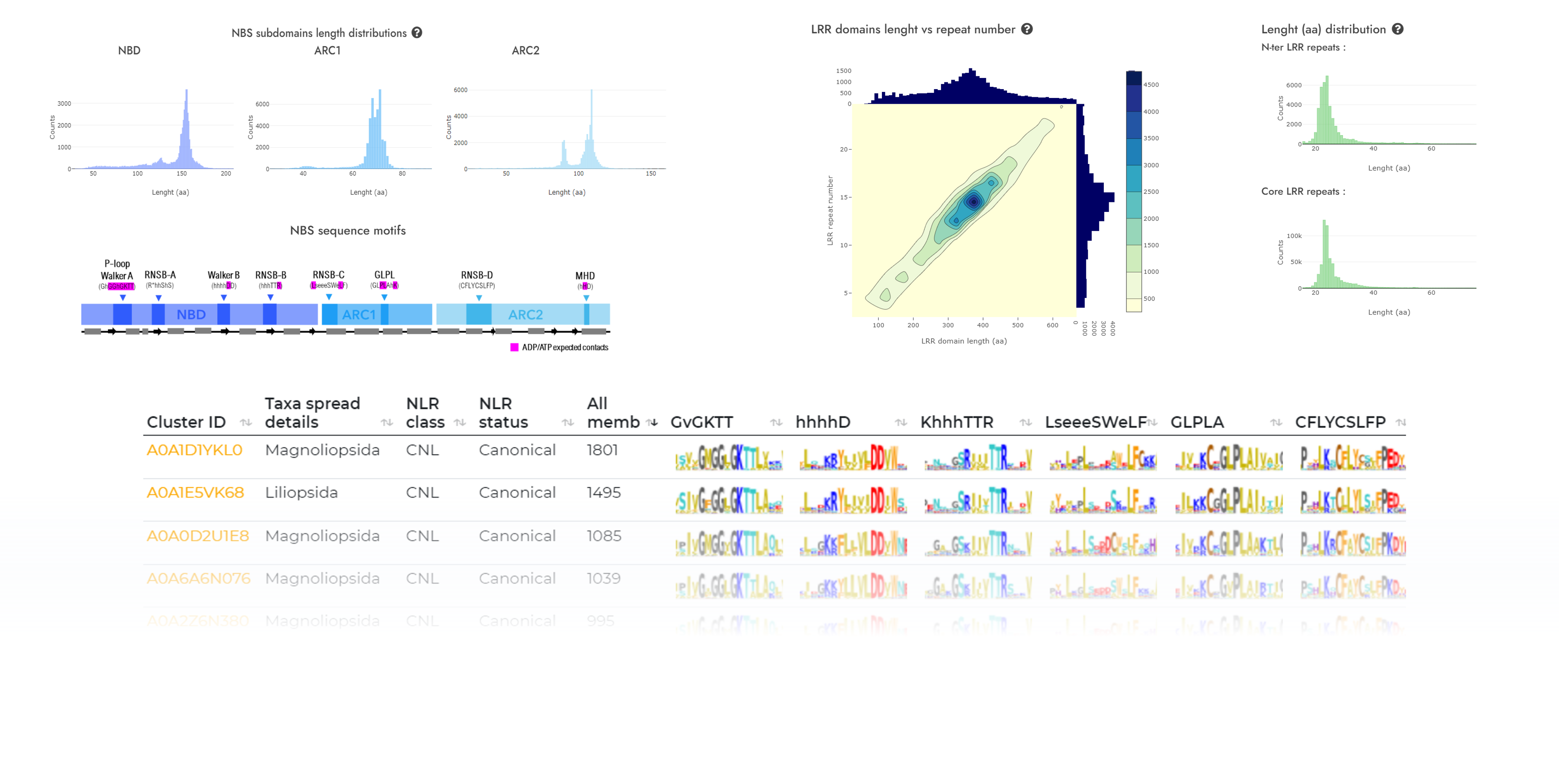
Explore plant NLR landscape
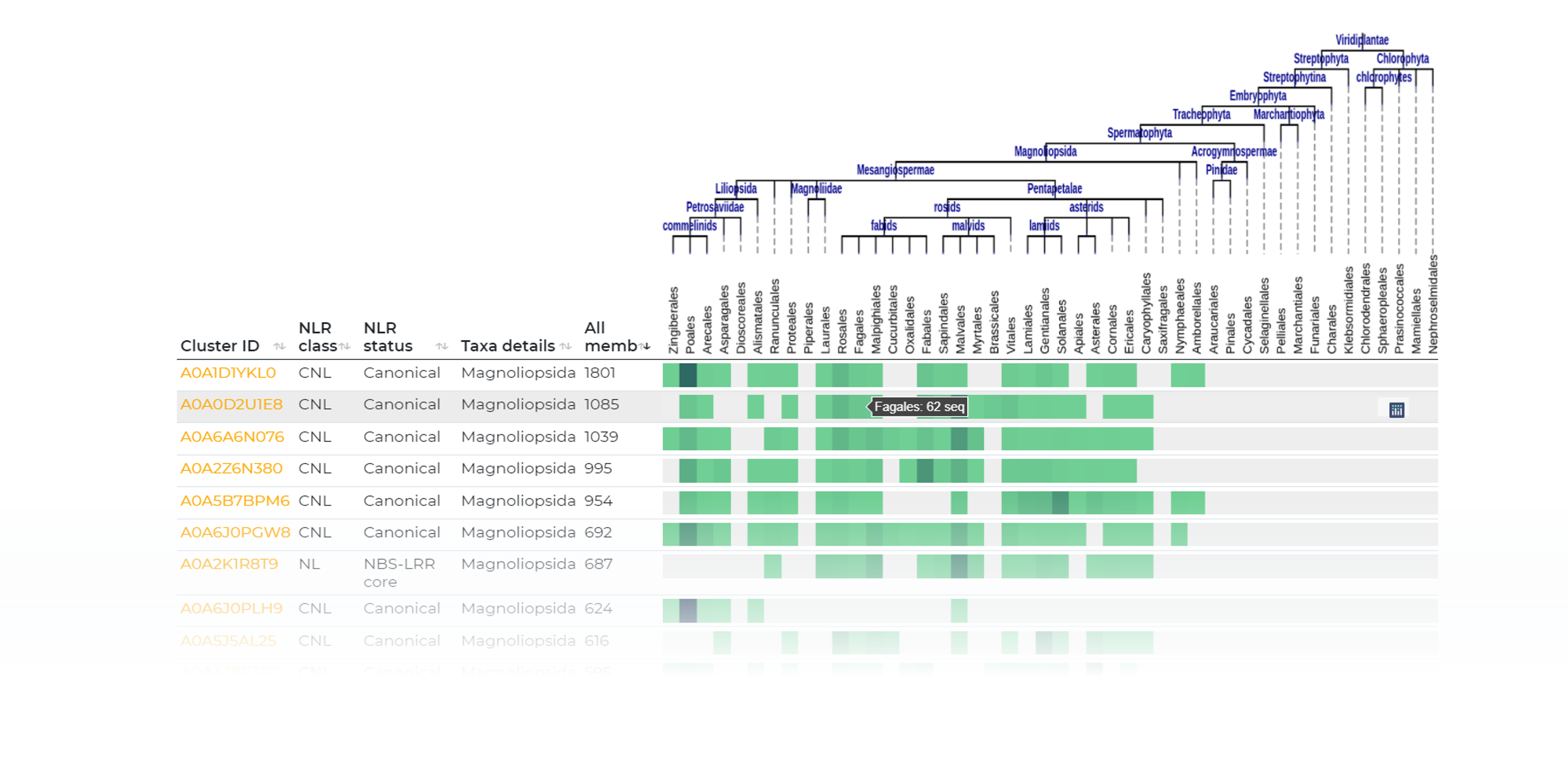
Taxa spread
Explore clusters based on taxonomic spread: from order-specific to phylum-spread at different homology cutoffs.
Bottom-up
Start from your sequence of interest
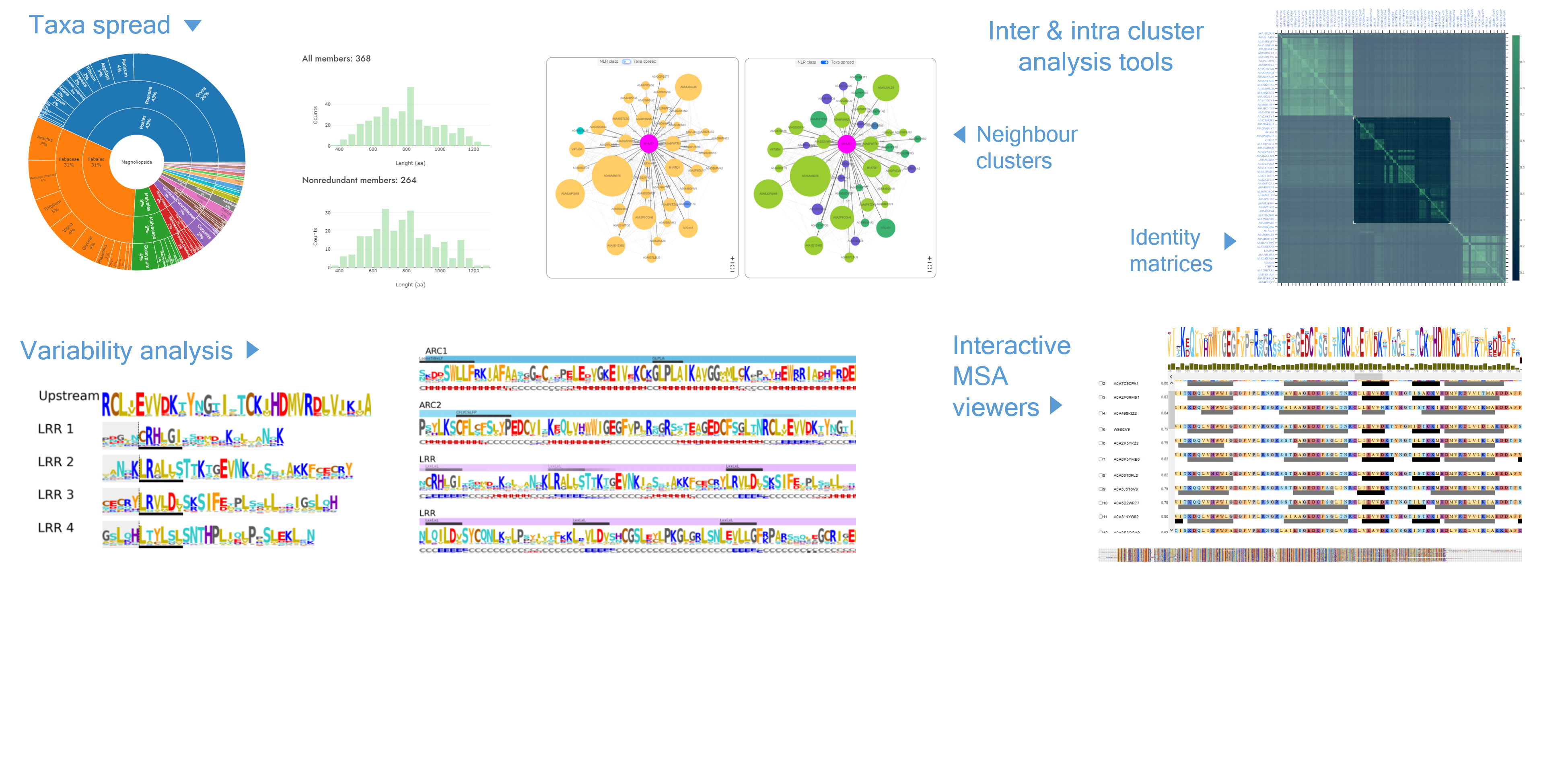
Locate your sequence
View outline sequence info, compare domain and motif annotations from multiple sources, examine predefined clusters containing your sequence.
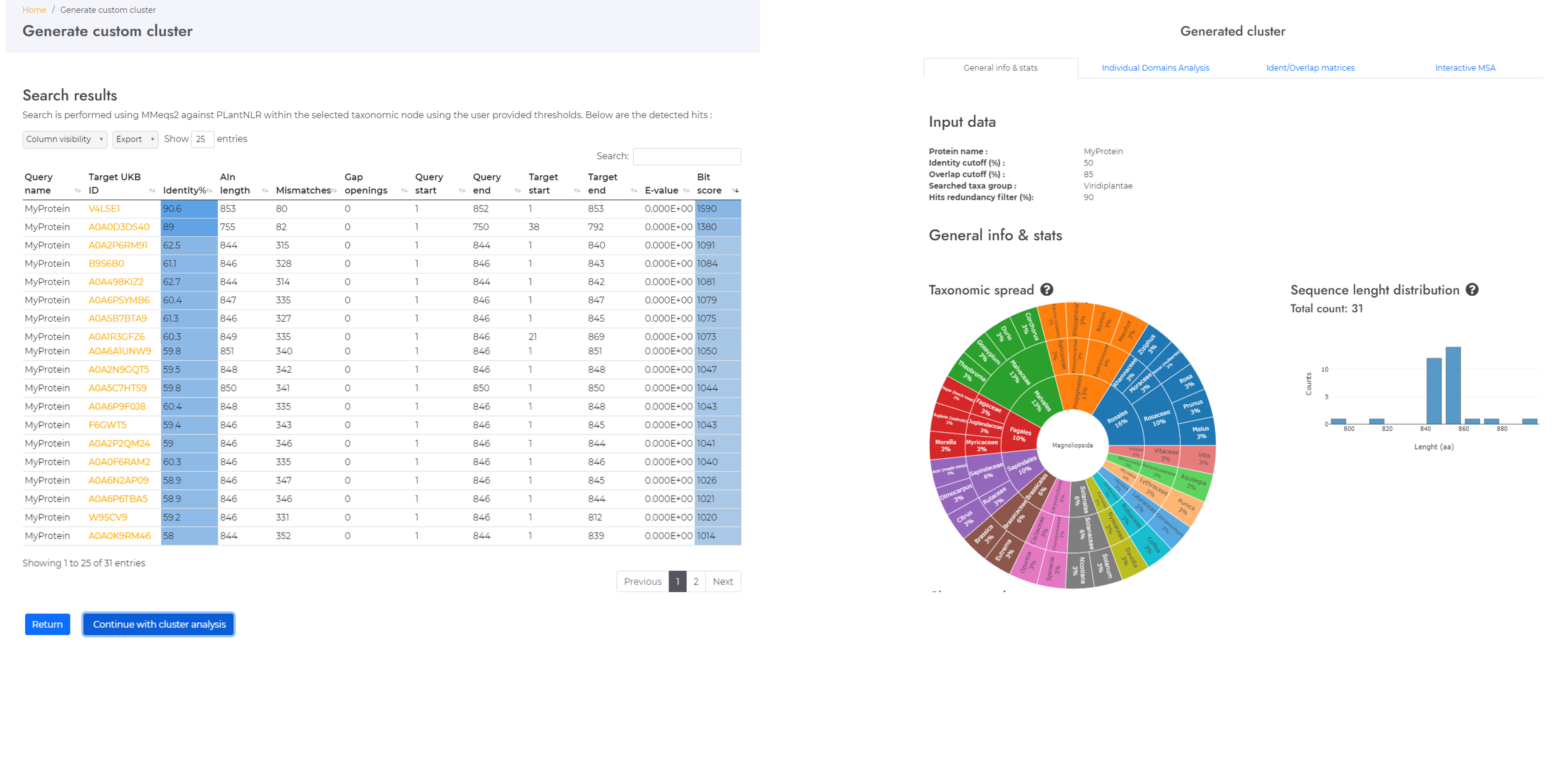
Generate custom clusters
Locate your sequence in the NLR landscape. Find neighbors, generate custom clusters centered around your sequence and perform analyzes using the NLRscape tools.
News
News regarding feature updates will be listed below:
-
11.08.2022
New updates include synoptic available data mapped on the amino acid sequence in sequence view page, containing
- - Mapped domain annotations from NLRscape, InterPro and Uniprot databases
- - Sequence conflicts, mutagenesis and variant data from Uniprot.
- - Secondary structure elements (currently only for experimental data)
-
15.12.2021
The beta version of the website is now released, new available features include :
- - Custom cluster generation feature.
- - Cluster map visualisation.
- - Cluster page: Neighbour clusters interactive graphs.
- - MSAviewer browser incompatibilities were solved.
-
05.06.2021
An initial version of the website is available at the moment, but only a fraction of the intended functionalities are integrated. By December 2021, a beta version of the website will be released. Prior to this, some functionalities will be on and off while the website is being finalized and parts of the data might be subjected to changes or updates.